Knowing the cellular profile of your kinase inhibitor is crucial
In 2018, we published a comparison between inhibitor profiles across the kinome generated using a biochemical in vitro assay or the cellular NanoBRETTM target engagement assay (Vasta, et al (2018)). We found the profiles for dasatinib and crizotinib differed tremendously!
What makes both assay formats different? In vitro assays frequently use only the catalytic domain of a purified enzyme as an input to determine enzyme activity. These proteins, that are most often expressed in bacterial cells or insect cells, are different from human expressed proteins e.g. based on their post-translational modifications. In the NanoBRETTM assay, we are using human HEK293T cells as an expression system for our targets as a full-length protein spanning not only the catalytic domain but also different domains that may be involved in scaffolding or protein structure and activity – thereby also creating human post-translational modification. In NanoBRETTM, we also take the cell membrane into account which your compounds need to pass in order to engage their target. In living cells, we also find endogenous levels of natural binding partners of our targets, e.g. Adenosine Triphosphate (ATP) at high concentrations of approximately 2 mM that will impact small molecule binding.
In our study from 2018, we found the individual protein kinases KM for ATP to be the reason for the differing inhibitor profiles (and subsequently a shift in their respective EC50s compared to in vitro IC50s) – making clear that testing your small molecule in living cells is absolutely necessary!
CELLinib128: Determine your kinase inhibitor’s profile using a representative kinome profiling for small molecules
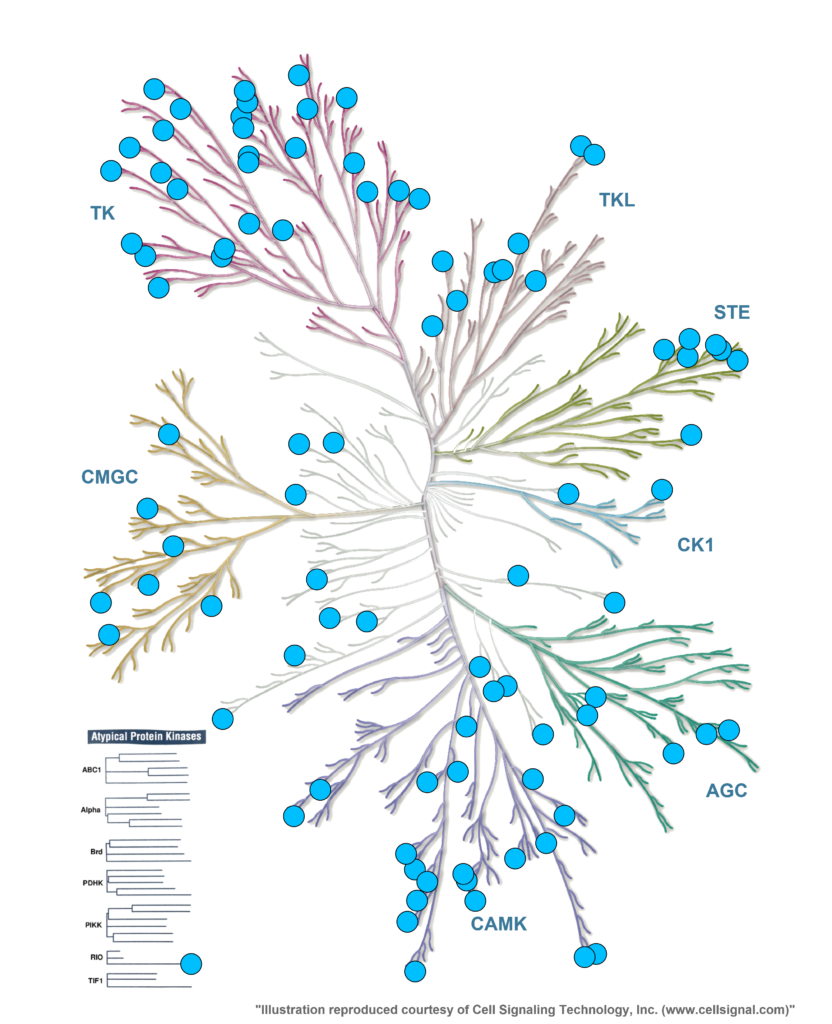
Screen of 128 representative protein kinases as a 1-shot technical duplicate assay using NanoBRET
- Assess your small molecules binding profile across a representative section of the human kinome
- Use human expressed full-length protein kinases with human post-translational modifications
- Determine target engagement in living cells (HEK293T)
- Assess binding under physiological conditions – with cellular concentrations of ATP and natural binding partners
CELLinib96 - representative panel
Test Molecule requirements
- 100 µL 1000x of your desired test concentration in 100% DMSO
- Test molecule needs to be soluble – we will not attempt to solubilize your molecule but test the stock as is
- Test molecule cytotoxicity may affect the assay outcome (because cell tox will lead to lower luciferase signal and hence lower assay performance) – we will test your sample as is
- Test molecule absorbtion at 450 nm will affect the assay outcome (less luciferase signal gets transfered for BRET) – we will test your sample as is
- Test molecule emission at 610 nm (after 450 nm excitation) will affect the assay outcome (artificially higher signal on the tracer channel) – we will test your sample as is
Fell free to contact us if you have any further questions!
